Developmental EEG Data
Contents
Developmental EEG Data¶
Analyze EEG data.
Dataset Details¶
Words, words, words.
from pathlib import Path
import numpy as np
import matplotlib.pyplot as plt
from fooof import FOOOF
from fooof.utils import trim_spectrum
from neurodsp.utils import create_times
from neurodsp.spectral import compute_spectrum
from neurodsp.plts import plot_time_series, plot_power_spectra
from bootstrap import bootstrap_corr, bootstrap_diff
# Import custom code
import sys
sys.path.append(str(Path('..').resolve()))
from apm.run import run_measures
from apm.plts import plot_dots
from apm.utils import format_corr
from apm.methods import specparam, irasa, dfa, hurst, hjorth_complexity, lempelziv, app_entropy
Settings¶
# Data path
data_path = Path('/Users/tom/Documents/Data/VoytekLab/ExtractedSubsets/childmind/')
# Define data information
fs = 500
n_seconds = 30
# Define times vector for data
times = create_times(n_seconds + 1/fs, fs)
Load Data¶
# Load data files
ages = np.load(data_path / 'ages.npy')
data = np.load(data_path / 'data.npy')
# Check the number of subjects
n_subjs = data.shape[0]
assert n_subjs == len(ages)
print('Number of subjects: {}'.format(n_subjs))
Number of subjects: 111
Data Checks¶
ind = 0
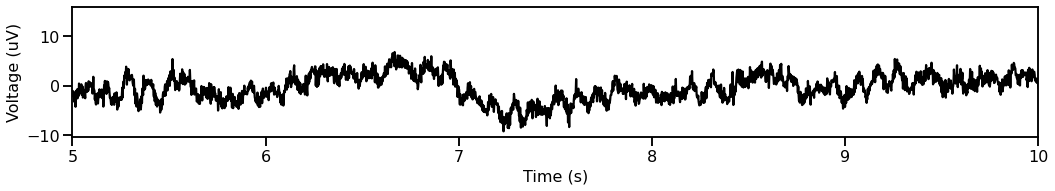
# Plot a segment of time series data
plot_time_series(times, data[ind, :], xlim=[5, 10])
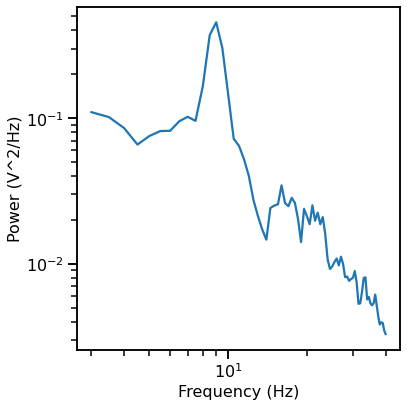
# Compute a power spectrum of an example
freqs, powers = compute_spectrum(data[ind, :], fs, nperseg=2*fs, noverlap=fs)
plot_power_spectra(*trim_spectrum(freqs, powers, [3, 40]))
Compare Methods¶
This is a replication of the
# Initialize specparam model
fm = FOOOF(verbose=False)
# General method settings
f_range = [3, 40]
# Define measures to apply
measures = {
specparam : {'fs' : fs, 'f_range' : f_range},
irasa : {'fs' : fs, 'f_range' : f_range},
hurst : {'fs' : fs},
dfa : {'fs' : fs},
lempelziv : {},
hjorth_complexity : {},
app_entropy : {}
}
# Compute measures of interest on the data
outputs = run_measures(data, measures)
Compare Measures¶
Compare Exponent Measures¶
plot_dots(outputs['specparam'], outputs['irasa'],
xlabel='Aperiodic Exponent (SP)', ylabel='Aperiodic Exponent (SP)')
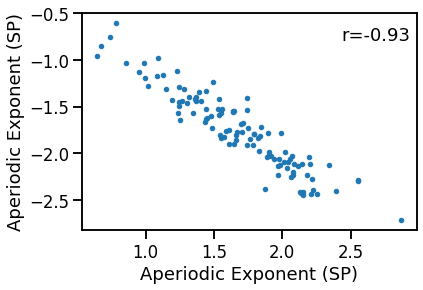
sp_ir_corrs = bootstrap_corr(outputs['specparam'], outputs['irasa'])
print(' SP & IR: ', format_corr(*sp_ir_corrs))
SP & IR: r=-0.932 CI[-0.953, -0.895], p=0.000
Compare exponent to LZ Complexity¶
plot_dots(outputs['specparam'], outputs['lempelziv'],
xlabel='Aperiodic Exponent', ylabel='LZ Complexity')
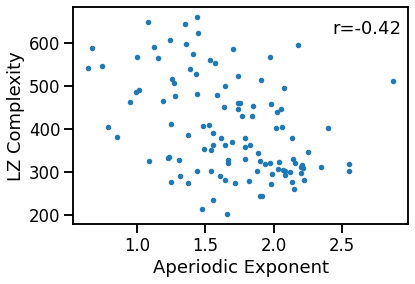
sp_lzc_corrs = bootstrap_corr(outputs['specparam'], outputs['lempelziv'])
print(' EXP & LZC: ', format_corr(*sp_lzc_corrs))
EXP & LZC: r=-0.422 CI[-0.567, -0.252], p=0.000
Compare exponent to Entropy¶
plot_dots(outputs['specparam'], outputs['app_entropy'],
xlabel='Aperiodic Exponent', ylabel='Entropy')
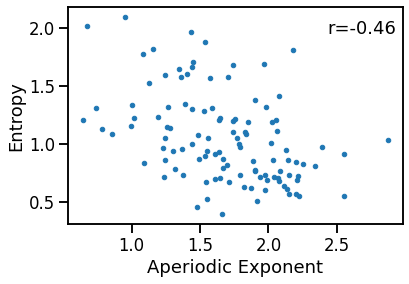
sp_ent_corrs = bootstrap_corr(outputs['specparam'], outputs['app_entropy'])
print(' SP & IR: ', format_corr(*sp_ent_corrs))
SP & IR: r=-0.463 CI[-0.598, -0.307], p=0.000
Check Age Relationship¶
Compare age to exponent¶
_, axes = plt.subplots(1, 2, figsize=(14, 5))
plot_dots(ages, outputs['specparam'], ax=axes[0],
xlabel='Age', ylabel='Aperiodic Exponent')
plot_dots(ages, -1 * outputs['irasa'], ax=axes[1],
xlabel='Age', ylabel='Aperiodic Exponent')
plt.subplots_adjust(wspace=0.25)
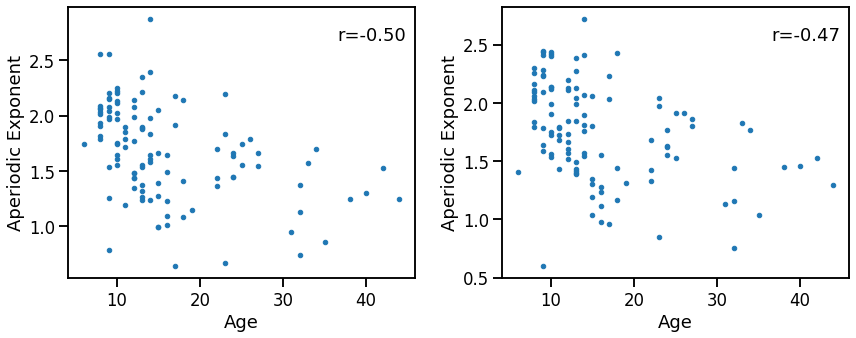
sp_age_corrs = bootstrap_corr(ages, outputs['specparam'])
print(' SPEXP & age: ', format_corr(*sp_age_corrs))
SPEXP & age: r=-0.502 CI[-0.624, -0.355], p=0.000
sp_age_corrs = bootstrap_corr(ages, outputs['irasa'])
print(' IREXP & age: ', format_corr(*sp_age_corrs))
IREXP & age: r=+0.469 CI[+0.314, +0.596], p=0.000
corr_diff = bootstrap_diff(ages, outputs['specparam'], -1 * outputs['irasa'])
print(' SP & IR age diff: ', format_corr(*corr_diff))
SP & IR age diff: r=-0.034 CI[-0.121, +0.044], p=0.403
Compare age to LZ Complexity¶
plot_dots(ages, outputs['lempelziv'],
xlabel='Age', ylabel='Lempel-Ziv Complexity')
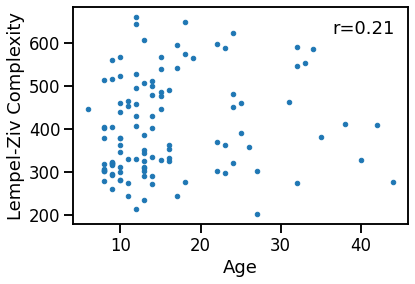
lcz_age_corrs = bootstrap_corr(ages, outputs['lempelziv'])
print(' SP & age: ', format_corr(*lcz_age_corrs))
SP & age: r=+0.213 CI[+0.032, +0.382], p=0.025
Compare age to Hjorth Complexity¶
plot_dots(ages, outputs['hjorth_complexity'],
xlabel='Age', ylabel='Hjorth Complexity')
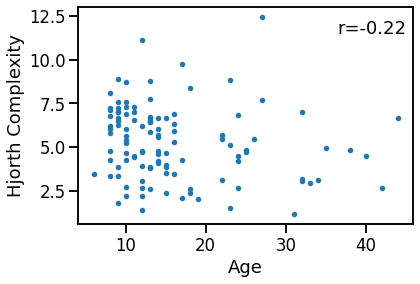
hcx_age_corrs = bootstrap_corr(ages, outputs['hjorth_complexity'])
print(' SP & age: ', format_corr(*hcx_age_corrs))
SP & age: r=-0.218 CI[-0.399, -0.035], p=0.021
Compare age to Entropy¶
plot_dots(ages, outputs['app_entropy'],
xlabel='Age', ylabel='Entropy')
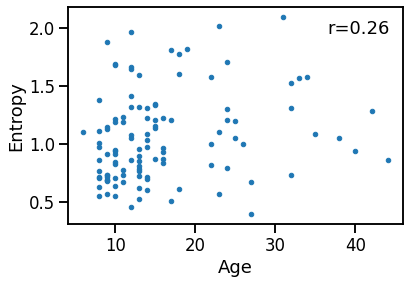
ent_age_corrs = bootstrap_corr(ages, outputs['app_entropy'])
print(' SP & age: ', format_corr(*ent_age_corrs))
SP & age: r=+0.260 CI[+0.084, +0.429], p=0.006
Conclusions¶
Words, words, words.