Complexity Measures
Contents
Complexity Measures¶
This notebooks runs complexity metrics on time series signals.
This notebook includes:
Hjorth parameters
Lempel-Ziv complexity
Fractal dimension measures
Note that while entropy measures can be considered as mesaured of complexity, they are considered in a separate notebook.
import numpy as np
import matplotlib.pyplot as plt
from neurodsp.sim import sim_powerlaw, sim_synaptic_current, sim_combined
from neurodsp.utils import set_random_seed
from neurodsp.aperiodic.conversions import convert_exponent_hfd
from antropy import lziv_complexity
# Import custom code
import sys; from pathlib import Path
sys.path.append(str(Path('..').resolve()))
from apm.run import run_sims
from apm.methods import (hjorth_activity, hjorth_mobility, hjorth_complexity,
lempelziv, higuchi_fd, petrosian_fd, katz_fd)
from apm.methods.settings import LZ_PARAMS
from apm.plts import plot_lines
from apm.plts.settings import AP_COL, COMB_COL
from apm.sim.settings import SIM_PARAMS_AP, SIM_PARAMS_COMB, EXPS, FREQS, POWERS, N_SIMS, EXP
from apm.sim.examples import SIG_AP, SIG_KN, SIG_OSC, SIG_COMB
Hjorth Parameters¶
Hjorth parameters are statistical properties that can be used to characterize EEG data.
The Hjorth measures are:
activity: represents the signal power, computed as the variance of the time series
mobility: represents the mean frequency of the power spectrum
complexity: represents the change in frequency
Compute Hjorth Parameters on Examples Signals¶
Note that the simulated signals are normalized to a variance of one, which is why the Hjorth activity is the same for all.
Powerlaw Signal¶
# Check the calculated Hjorth activity values across some example signals
print('Computed Hjorth activity:')
print(' powerlaw: \t {:1.4f}'.format(hjorth_activity(SIG_AP)))
print(' osc: \t {:6.4f}'.format(hjorth_activity(SIG_OSC)))
print(' combined: \t {:1.4f}'.format(hjorth_activity(SIG_COMB)))
print(' knee: \t {:1.4f}'.format(hjorth_activity(SIG_KN)))
Computed Hjorth activity:
powerlaw: 1.0000
osc: 1.0000
combined: 1.0000
knee: 1.0000
# Check the calculated Hjorth mobility values across some example signals
print('Computed Hjorth mobility:')
print(' powerlaw: \t {:1.4f}'.format(hjorth_mobility(SIG_AP)))
print(' osc: \t {:6.4f}'.format(hjorth_mobility(SIG_OSC)))
print(' combined: \t {:1.4f}'.format(hjorth_mobility(SIG_COMB)))
print(' knee: \t {:1.4f}'.format(hjorth_mobility(SIG_KN)))
Computed Hjorth mobility:
powerlaw: 0.1281
osc: 0.0628
combined: 0.2453
knee: 0.4362
# Check the calculated Hjorth mobility values across some example signals
print('Computed Hjorth complexity:')
print(' powerlaw: \t {:1.4f}'.format(hjorth_complexity(SIG_AP)))
print(' osc: \t {:6.4f}'.format(hjorth_complexity(SIG_OSC)))
print(' combined: \t {:1.4f}'.format(hjorth_complexity(SIG_COMB)))
print(' knee: \t {:1.4f}'.format(hjorth_complexity(SIG_KN)))
Computed Hjorth complexity:
powerlaw: 11.4881
osc: 1.0001
combined: 5.9377
knee: 3.3120
Hjorth Parameters: Aperiodic Variations¶
# Run a set of simulations, calculating Hjorth measures across exponents
outs = run_sims(sim_powerlaw, SIM_PARAMS_AP, hjorth_mobility, {},
'update_exp', EXPS, N_SIMS, var_func=np.std)
hjm_sims_exp, hjm_sims_exp_var = outs
outs = run_sims(sim_powerlaw, SIM_PARAMS_AP, hjorth_complexity, {},
'update_exp', EXPS, N_SIMS, var_func=np.std)
hjc_sims_exp, hjc_sims_exp_var = outs
# Run a set of simulations, calculating Hjorth measures across exponents with an oscillation
outs = run_sims(sim_combined, SIM_PARAMS_COMB, hjorth_mobility, {},
'update_comb_exp', EXPS, N_SIMS, var_func=np.std)
hjm_sims_comb, hjm_sims_comb_var = outs
outs = run_sims(sim_combined, SIM_PARAMS_COMB, hjorth_complexity, {},
'update_comb_exp', EXPS, N_SIMS, var_func=np.std)
hjc_sims_comb, hjc_sims_comb_var = outs
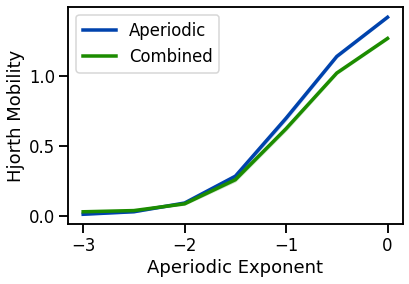
# Plot Hjorth mobility across exponents
plot_lines(EXPS, hjm_sims_exp, hjm_sims_exp_var, color=AP_COL, label='Aperiodic')
plot_lines(EXPS, hjm_sims_comb, hjm_sims_comb_var, color=COMB_COL, label='Combined')
plot_lines(xlabel='Aperiodic Exponent', ylabel='Hjorth Mobility')
plot_lines(save_fig=SAVE_FIG, file_name='hjm_both', file_path='complexity')
# Plot Hjorth complexity across exponents with an oscillation present
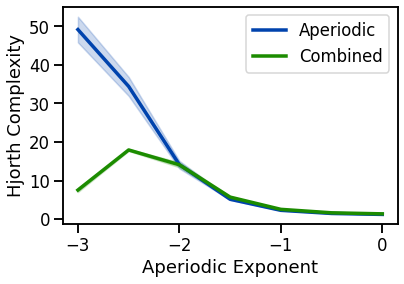
plot_lines(EXPS, hjc_sims_exp, hjc_sims_exp_var, color=AP_COL, label='Aperiodic')
plot_lines(EXPS, hjc_sims_comb, hjc_sims_comb_var, color=COMB_COL, label='Combined')
plot_lines(xlabel='Aperiodic Exponent', ylabel='Hjorth Complexity')
plot_lines(save_fig=SAVE_FIG, file_name='hjc_both', file_path='complexity')
Hjorth Parameters: Periodic Variations¶
# Run a set of simulations, calculating fractal dimension across oscillation frequency
outs = run_sims(sim_combined, SIM_PARAMS_COMB, hjorth_mobility, {},
'update_freq', FREQS, N_SIMS, var_func=np.std)
hjm_sims_freq, hjm_sims_freq_var = outs
outs = run_sims(sim_combined, SIM_PARAMS_COMB, hjorth_complexity, {},
'update_freq', FREQS, N_SIMS, var_func=np.std)
hjc_sims_freq, hjc_sims_freq_var = outs
# Run a set of simulations, calculating fractal dimension across oscillation power
outs = run_sims(sim_combined, SIM_PARAMS_COMB, hjorth_mobility, {},
'update_pow', POWERS, N_SIMS, var_func=np.std)
hjm_sims_pow, hjm_sims_pow_var = outs
outs = run_sims(sim_combined, SIM_PARAMS_COMB, hjorth_complexity, {},
'update_pow', POWERS, N_SIMS, var_func=np.std)
hjc_sims_pow, hjc_sims_pow_var = outs
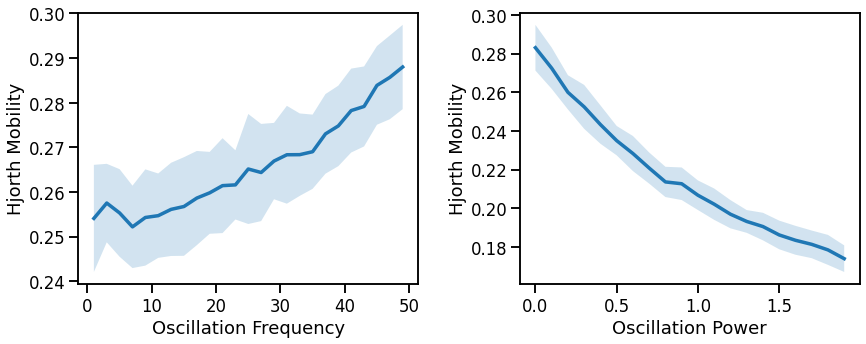
# Plot effect of oscillation variation on Hjorth mobility
fig, axes = plt.subplots(1, 2, figsize=(14, 5))
plot_lines(FREQS, hjm_sims_freq, hjm_sims_freq_var, ax=axes[0])
plot_lines(xlabel='Oscillation Frequency', ylabel='Hjorth Mobility', ax=axes[0])
plot_lines(POWERS, hjm_sims_pow, hjm_sims_pow_var, ax=axes[1])
plot_lines(xlabel='Oscillation Power', ylabel='Hjorth Mobility', ax=axes[1])
plt.subplots_adjust(wspace=0.3)
plot_lines(save_fig=SAVE_FIG, file_name='hjm_osc_both', file_path='complexity')
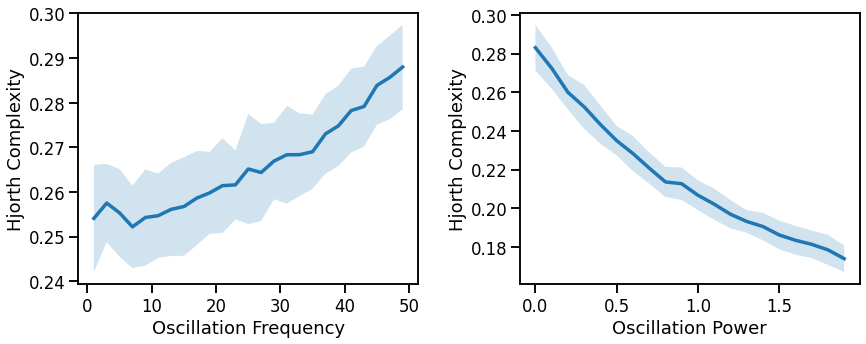
# Plot effect of oscillation variation on Hjorth complexity
fig, axes = plt.subplots(1, 2, figsize=(14, 5))
plot_lines(FREQS, hjm_sims_freq, hjm_sims_freq_var, ax=axes[0])
plot_lines(xlabel='Oscillation Frequency', ylabel='Hjorth Complexity', ax=axes[0])
plot_lines(POWERS, hjm_sims_pow, hjm_sims_pow_var, ax=axes[1])
plot_lines(xlabel='Oscillation Power', ylabel='Hjorth Complexity', ax=axes[1])
plt.subplots_adjust(wspace=0.3)
plot_lines(save_fig=SAVE_FIG, file_name='hjc_osc_both', file_path='complexity')
Lempel-Ziv Complexity¶
Lempel-Ziv (LZ) complexity is a measure of the complexity of a time series that is sometimes applied to EEG data.
Note that the LZ algorithm is defined for binary sequences.
To apply this measure to neural time series, we binarize the signal by calculating whether each point is above or below the median of the time series.
# Check Lempel-Ziv complexity parameters
LZ_PARAMS
{'normalize': False}
# Example of applying LZ complexity to a binarized signal
bin_sig = np.array(SIG_AP > np.median(SIG_AP)).astype(int)
lziv_complexity(bin_sig)
395
Calculating Lempel-Ziv Complexity on Example Signals¶
# Check Lempel-Ziv complexity measures on example signals
print('Lempel-Ziv complexity:')
print(' powerlaw: \t {:5d}'.format(lempelziv(SIG_AP, **LZ_PARAMS)))
print(' osc: \t {:5d}'.format(lempelziv(SIG_OSC, **LZ_PARAMS)))
print(' combined: \t {:5d}'.format(lempelziv(SIG_COMB, **LZ_PARAMS)))
print(' knee: \t {:5d}'.format(lempelziv(SIG_KN, **LZ_PARAMS)))
Lempel-Ziv complexity:
powerlaw: 395
osc: 5
combined: 708
knee: 1115
Aperiodic Variations¶
# Run a set of simulations, calculating LZ complexity across exponents
outs = run_sims(sim_powerlaw, SIM_PARAMS_AP, lempelziv, LZ_PARAMS,
'update_exp', EXPS, N_SIMS, var_func=np.std)
lz_sims_exp, lz_sims_exp_var = outs
# Run a set of simulations, calculating LZ complexity across exponents with an oscillation
outs = run_sims(sim_combined, SIM_PARAMS_COMB, lempelziv, LZ_PARAMS,
'update_comb_exp', EXPS, N_SIMS, var_func=np.std)
lz_sims_comb, lz_sims_comb_var = outs
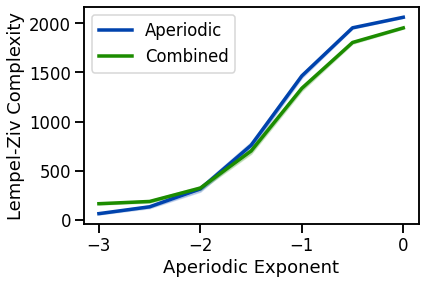
# Plot LZ complexity measures
plot_lines(EXPS, lz_sims_exp, lz_sims_exp_var, color=AP_COL, label='Aperiodic')
plot_lines(EXPS, lz_sims_comb, lz_sims_comb_var, color=COMB_COL, label='Combined')
plot_lines(xlabel='Aperiodic Exponent', ylabel='Lempel-Ziv Complexity')
plot_lines(save_fig=SAVE_FIG, file_name='lz_both', file_path='complexity')
Periodic Variations¶
# Run a set of simulations, calculating LZ complexity across oscillation frequency
outs = run_sims(sim_combined, SIM_PARAMS_COMB, lempelziv, LZ_PARAMS,
'update_freq', FREQS, N_SIMS, var_func=np.std)
lz_sims_freq, lz_sims_freq_var = outs
# Run a set of simulations, calculating fractal dimension across oscillation power
outs = run_sims(sim_combined, SIM_PARAMS_COMB, lempelziv, LZ_PARAMS,
'update_pow', POWERS, N_SIMS, var_func=np.std)
lz_sims_pow, lz_sims_pow_var = outs
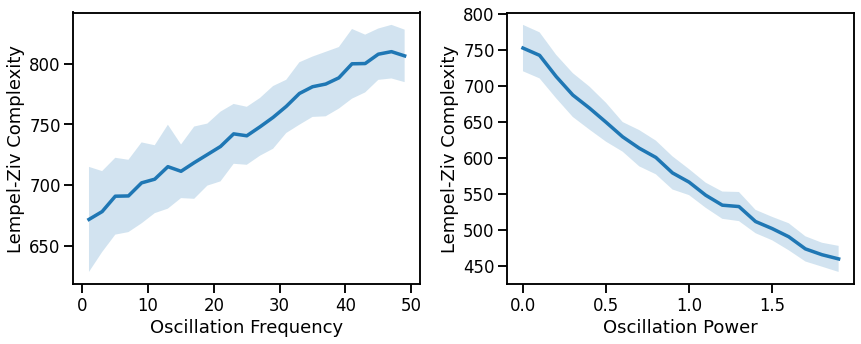
# Plot effect of oscillation variation on LZ complexity
fig, axes = plt.subplots(1, 2, figsize=(14, 5))
plot_lines(FREQS, lz_sims_freq, lz_sims_freq_var, ax=axes[0])
plot_lines(xlabel='Oscillation Frequency', ylabel='Lempel-Ziv Complexity', ax=axes[0])
plot_lines(POWERS, lz_sims_pow, lz_sims_pow_var, ax=axes[1])
plot_lines(xlabel='Oscillation Power', ylabel='Lempel-Ziv Complexity', ax=axes[1])
plt.subplots_adjust(wspace=0.25)
plot_lines(save_fig=SAVE_FIG, file_name='lz_osc_both', file_path='complexity')
Fractal Dimensions¶
Calculate measures of fractal dimension on time series.
Fractal dimension measures:
Higuchi Fractal Dimension
Petrosian Fractal Dimension
Katz Fractal Dimension
Calculating Fractal Dimension on Example Signals¶
# Check Higuchi fractal dimension measures on example signals
print('Higuchi fractal dimension:')
print(' powerlaw: \t {:1.4f} \t {:1.2f}'.format(higuchi_fd(SIG_AP), convert_exponent_hfd(-EXP)))
print(' osc: \t {:1.4f}'.format(higuchi_fd(SIG_OSC)))
print(' combined: \t {:1.4f}'.format(higuchi_fd(SIG_COMB)))
print(' knee: \t {:1.4f}'.format(higuchi_fd(SIG_KN)))
Higuchi fractal dimension:
powerlaw: 1.6557 1.75
osc: 1.0069
combined: 1.6112
knee: 1.5784
# Check Katz fractal dimension measures on example signals
print('Katz fractal dimension:')
print(' powerlaw: \t {:1.4f}'.format(katz_fd(SIG_AP)))
print(' osc: \t {:1.4f}'.format(katz_fd(SIG_OSC)))
print(' combined: \t {:1.4f}'.format(katz_fd(SIG_COMB)))
print(' knee: \t {:1.4f}'.format(katz_fd(SIG_KN)))
Katz fractal dimension:
powerlaw: 2.8828
osc: 3.2026
combined: 3.1405
knee: 3.8000
# Check Petrosian fractal dimension measures on example signals
print('Petrosian fractal dimension:')
print(' powerlaw: \t {:1.4f}'.format(petrosian_fd(SIG_AP)))
print(' osc: \t {:1.4f}'.format(petrosian_fd(SIG_OSC)))
print(' combined: \t {:1.4f}'.format(petrosian_fd(SIG_COMB)))
print(' knee: \t {:1.4f}'.format(petrosian_fd(SIG_KN)))
Petrosian fractal dimension:
powerlaw: 1.0189
osc: 1.0008
combined: 1.0185
knee: 1.0185
Fractal Dimension: Aperiodic Variations¶
# Run a set of simulations, calculating fractal dimension across exponents
outs = run_sims(sim_powerlaw, SIM_PARAMS_AP, higuchi_fd, {},
'update_exp', EXPS, N_SIMS, var_func=np.std)
hfd_sims_exp, hfd_sims_exp_var = outs
outs = run_sims(sim_powerlaw, SIM_PARAMS_AP, katz_fd, {},
'update_exp', EXPS, N_SIMS, var_func=np.std)
kfd_sims_exp, kfd_sims_exp_var = outs
outs = run_sims(sim_powerlaw, SIM_PARAMS_AP, petrosian_fd, {},
'update_exp', EXPS, N_SIMS, var_func=np.std)
pfd_sims_exp, pfd_sims_exp_var = outs
# Run a set of simulations, calculating fractal dimension across exponents with an oscillation
outs = run_sims(sim_combined, SIM_PARAMS_COMB, higuchi_fd, {},
'update_comb_exp', EXPS, N_SIMS, var_func=np.std)
hfd_sims_comb, hfd_sims_comb_var = outs
outs = run_sims(sim_combined, SIM_PARAMS_COMB, katz_fd, {},
'update_comb_exp', EXPS, N_SIMS, var_func=np.std)
kfd_sims_comb, kfd_sims_comb_var = outs
outs = run_sims(sim_combined, SIM_PARAMS_COMB, petrosian_fd, {},
'update_comb_exp', EXPS, N_SIMS, var_func=np.std)
pfd_sims_comb, pfd_sims_comb_var = outs
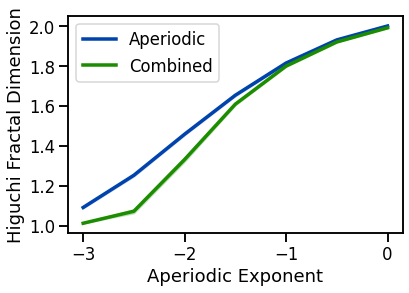
# Plot Higuchi fractal dimension measures
plot_lines(EXPS, hfd_sims_exp, hfd_sims_exp_var, color=AP_COL, label='Aperiodic')
plot_lines(EXPS, hfd_sims_comb, hfd_sims_comb_var, color=COMB_COL, label='Combined')
plot_lines(xlabel='Aperiodic Exponent', ylabel='Higuchi Fractal Dimension')
plot_lines(save_fig=SAVE_FIG, file_name='hfd_both', file_path='complexity')
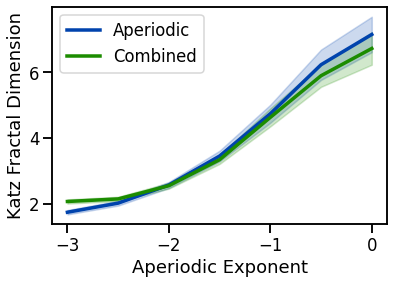
# Plot Katz fractal dimension measures
plot_lines(EXPS, kfd_sims_exp, kfd_sims_exp_var, color=AP_COL, label='Aperiodic')
plot_lines(EXPS, kfd_sims_comb, kfd_sims_comb_var, color=COMB_COL, label='Combined')
plot_lines(xlabel='Aperiodic Exponent', ylabel='Katz Fractal Dimension')
plot_lines(save_fig=SAVE_FIG, file_name='kfd_both', file_path='complexity')
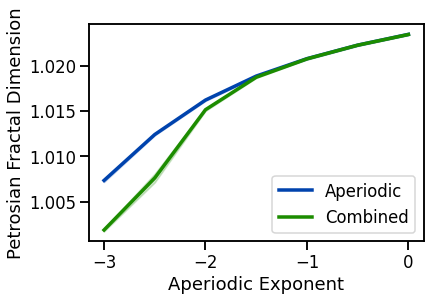
# Plot Petrosian fractal dimension measures
plot_lines(EXPS, pfd_sims_exp, pfd_sims_exp_var, color=AP_COL, label='Aperiodic')
plot_lines(EXPS, pfd_sims_comb, pfd_sims_comb_var, color=COMB_COL, label='Combined')
plot_lines(xlabel='Aperiodic Exponent', ylabel='Petrosian Fractal Dimension')
plot_lines(save_fig=SAVE_FIG, file_name='pfd_both', file_path='complexity')
Fractal Dimension: Periodic Variations¶
# Run a set of simulations, calculating fractal dimension across oscillation frequency
outs = run_sims(sim_combined, SIM_PARAMS_COMB, higuchi_fd, {},
'update_freq', FREQS, N_SIMS, var_func=np.std)
hfd_sims_freq, hfd_sims_freq_var = outs
outs = run_sims(sim_combined, SIM_PARAMS_COMB, katz_fd, {},
'update_freq', FREQS, N_SIMS, var_func=np.std)
kfd_sims_freq, kfd_sims_freq_var = outs
outs = run_sims(sim_combined, SIM_PARAMS_COMB, petrosian_fd, {},
'update_freq', FREQS, N_SIMS, var_func=np.std)
pfd_sims_freq, pfd_sims_freq_var = outs
# Run a set of simulations, calculating fractal dimension across oscillation power
outs = run_sims(sim_combined, SIM_PARAMS_COMB, higuchi_fd, {},
'update_pow', POWERS, N_SIMS, var_func=np.std)
hfd_sims_pow, hfd_sims_pow_var = outs
outs = run_sims(sim_combined, SIM_PARAMS_COMB, katz_fd, {},
'update_pow', POWERS, N_SIMS, var_func=np.std)
kfd_sims_pow, kfd_sims_pow_var = outs
outs = run_sims(sim_combined, SIM_PARAMS_COMB, petrosian_fd, {},
'update_pow', POWERS, N_SIMS, var_func=np.std)
pfd_sims_pow, pfd_sims_pow_var = outs
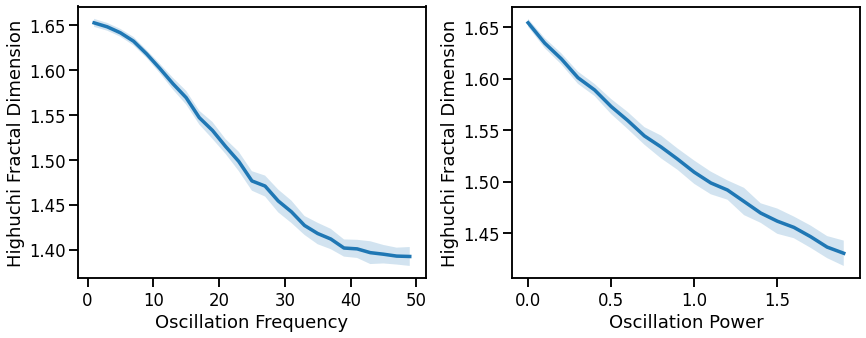
# Plot effect of oscillation variation on Higuchi fractal dimension
fig, axes = plt.subplots(1, 2, figsize=(14, 5))
plot_lines(FREQS, hfd_sims_freq, hfd_sims_freq_var, ax=axes[0])
plot_lines(xlabel='Oscillation Frequency', ylabel='Highuchi Fractal Dimension', ax=axes[0])
plot_lines(POWERS, hfd_sims_pow, hfd_sims_pow_var, ax=axes[1])
plot_lines(xlabel='Oscillation Power', ylabel='Highuchi Fractal Dimension', ax=axes[1])
plt.subplots_adjust(wspace=0.25)
plot_lines(save_fig=SAVE_FIG, file_name='hfd_osc_both', file_path='complexity')
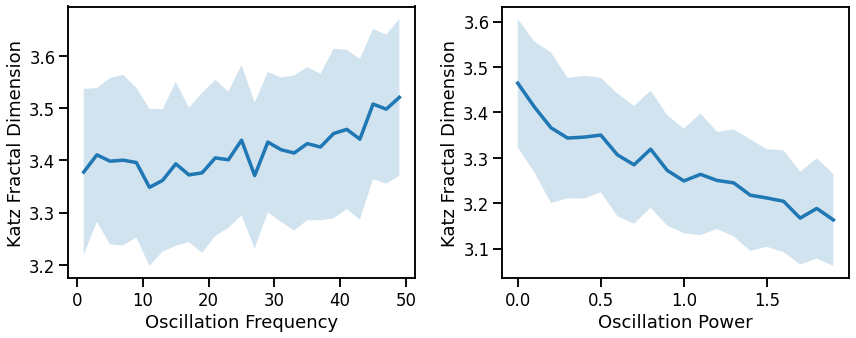
# Plot effect of oscillation variation on Katz fractal dimension
fig, axes = plt.subplots(1, 2, figsize=(14, 5))
plot_lines(FREQS, kfd_sims_freq, kfd_sims_freq_var, ax=axes[0])
plot_lines(xlabel='Oscillation Frequency', ylabel='Katz Fractal Dimension', ax=axes[0])
plot_lines(POWERS, kfd_sims_pow, kfd_sims_pow_var, ax=axes[1])
plot_lines(xlabel='Oscillation Power', ylabel='Katz Fractal Dimension', ax=axes[1])
plt.subplots_adjust(wspace=0.25)
plot_lines(save_fig=SAVE_FIG, file_name='kfd_osc_both', file_path='complexity')
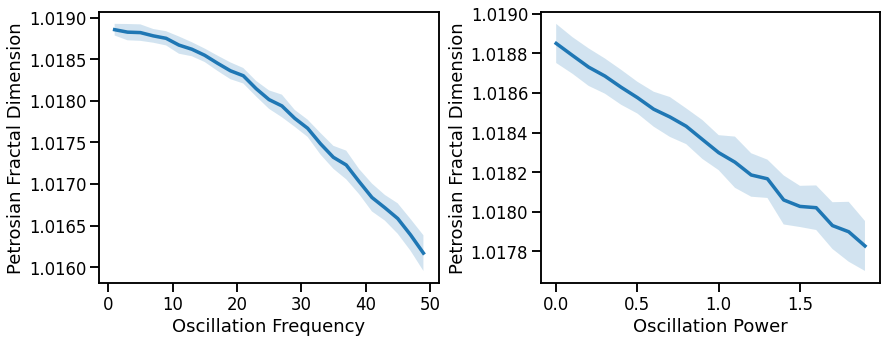
# Plot effect of oscillation variation on Petrosian fractal dimension
fig, axes = plt.subplots(1, 2, figsize=(14, 5))
plot_lines(FREQS, pfd_sims_freq, pfd_sims_freq_var, ax=axes[0])
plot_lines(xlabel='Oscillation Frequency', ylabel='Petrosian Fractal Dimension', ax=axes[0])
plot_lines(POWERS, pfd_sims_pow, pfd_sims_pow_var, ax=axes[1])
plot_lines(xlabel='Oscillation Power', ylabel='Petrosian Fractal Dimension', ax=axes[1])
plt.subplots_adjust(wspace=0.3)
plot_lines(save_fig=SAVE_FIG, file_name='pfd_osc_both', file_path='complexity')
Conclusions¶
Overall, we can see the following patterns in these simulations:
Hjorth Parameters:
mobility increases with decreasing aperiodic exponent (non-linearly) & increases with oscillation frequency & power
complexity decreases with decreasing aperiodic exponent (non-linearly) & decreases with oscillation frequency & power
Lempel-Ziv Complexity:
increases with decreasing aperiodic exponent, non-linearly
increases with increasing oscillation frequency, approximately linearly
decreases with increasing oscillation power, approximately linearly
Fractal dimension measures (general patterns, across all three):
increase with decreasing aperiodic exponent, non-linearly
decreases with increasing oscillations frequency or power, mixed patterns